Up to now, protein structure prediction and design problems have been mostly formulated assuming that proteins fold into a well-defined three-dimensional form. Nevertheless, there is an increasing corpus of work showing that Intrinsically Disordered Proteins/Regions (IDPs/IDRs) perform highly relevant biological tasks despite their lack of a permanent secondary or tertiary structure. Although substantial research efforts have been made in the past few years, much more remains to be done to understand and rationalize the relationships between sequence, structural properties and function for IDPs/IDRs.
In recent years, LAAS and CBS have successfully collaborated on the development of novel IDP modeling methods and tools, with biophysical validation. The first objective of the CORNFLEX project is to further improve our predictive modeling methods with the collaboration of IMT, and to extend them to address IDP/IDR design problems, based on an original formulation building on concepts and recent advances in statistics and artificial intelligence. With these new tools, we aim at deriving a deeper understanding of the relationships between the amino acid sequence and structural properties. This will enable us to anticipate structural perturbations exerted by sequence modifications and, finally, to design artificial (de novo) highly flexible proteins with tailored properties.
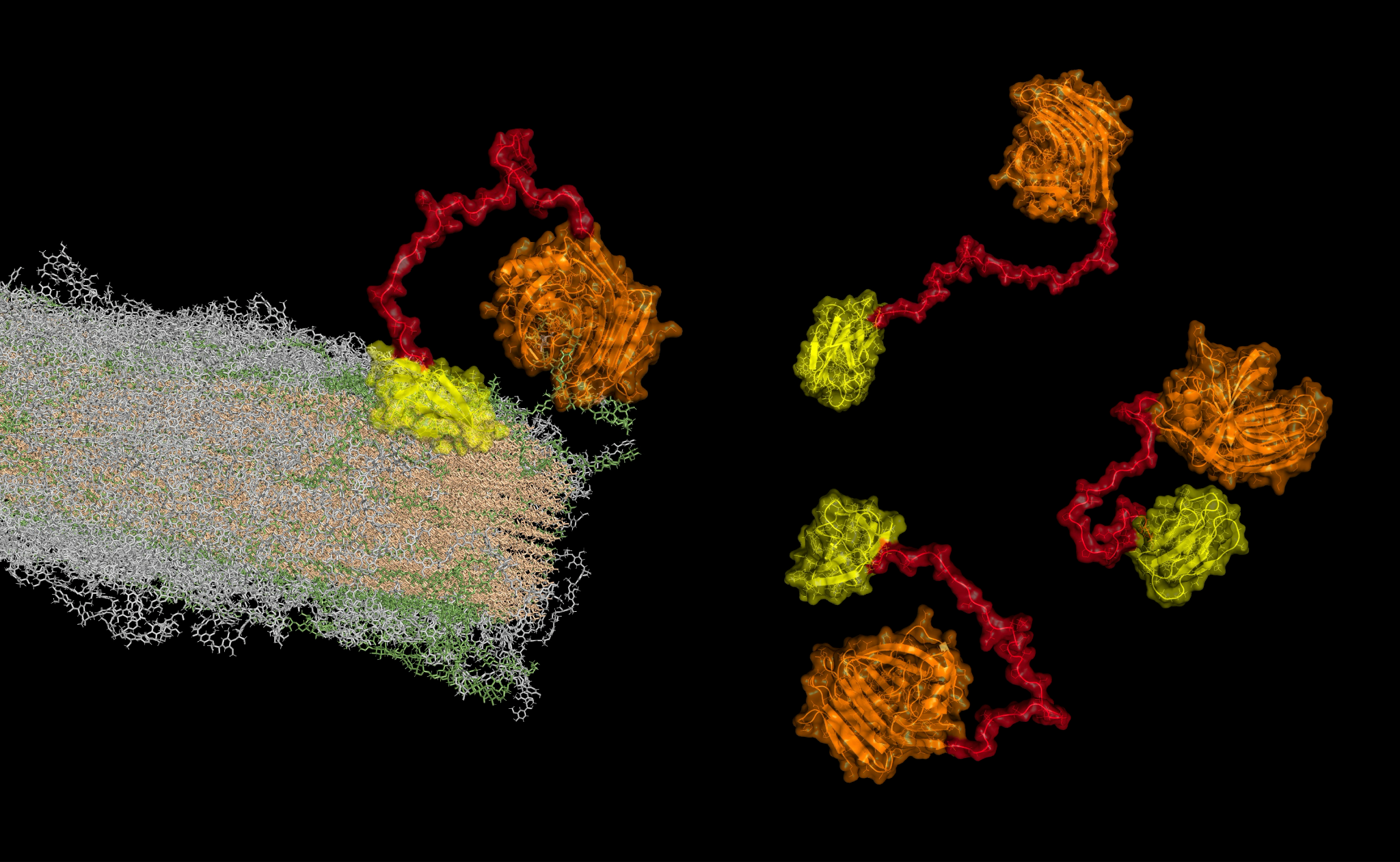
Although the developed methods aim to be general, we will focus on a particular proof-of-concept application: the design of flexible linkers in multi-modular enzymes in order to improve their catalytic activity toward the deconstruction of plant cell wall (PCW). For this, the consortium also involves two expert partners in enzyme engineering and plant biology: TBI and LRSV, respectively. PCW biomass has a vast potential as a renewable resource. Thus, the results of this project are aimed to bring methodological solutions to help building an environmentally sustainable economy.